Chapter 10
Atomistic and Particle-in-Cell Simulation
When motivating the Boltzmann equation it was argued that there are too
many particles for us to track them all, so we had to use a
distribution function approach. However, for some purposes, the number
of particles that need to be tracked can in fact be managable. In that
case, we can adopt a computational modelling approach that is called
generically Atomistic Simulation. In short, it involves time-advancing
individual classical particles obeying Newton's laws of motion (or in
some relativistic cases Einstein's extensions of them).
There are also situations where it is advantageous to solve Boltmann's
equation by a method that uses
pseudo-particles, each of them representative
of a large number of individual particles. These pseudo-particles are
modelled as if they were real particles. We'll come to this broader
topic in a later section, for now just think of real particles.
10.1 Atomistic Simulation
If the particles are literally atoms or molecules, then since we will
find it increasingly computationally difficult to track more than,
say, a few billion of them, the volume of material that can be
modelled is limited. A billion is 109 = 10003; so we could at most
model a three-dimensional crystal lattice roughly 1000 atoms on a
side. For a solid, that means the size of the region we can model is
perhaps 100 nm. Nanoscale. There's no way that we are
going to atomistically model a macroscopic (say 1 mm) piece of
material globally with forseeable computational capacity. Still, many
very interesting and important phenomena relating to solid defects,
atomic displacement due to energetic particle interactions, cracking,
surface behavior, and so on, occur on the 100 nm scale. Materials
behavior and design is a very important area for this direct atomic or
molecular simulation.
The timescale in atomic interactions ranges from, say, the time it
takes a gamma ray to cross a nucleus ∼ 10−23s to geological
times of ∼ 1014s. Modelling must choose a managable fraction of
this enormous range, because the particle time-steps must be shorter
than the fastest phenomenon to be modelled, and yet we can only afford
to compute a moderate number of steps, maybe routinely as many as
104, but not usually 106, and only heroically 108. Phenomena
outside our time-scale of choice have to be either irrelevant or
represented by simplified representations of their effects in our
modelling time-scale. Fig. 10.1 illustrates the
computationally feasible space and time-scale region (shaded) indicating the
approximate location of a few key phenomena.
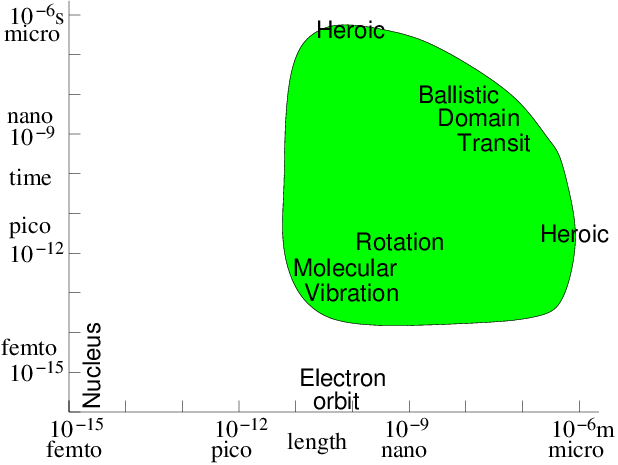
Figure 10.1: Approximate space and time scales for Molecular Dynamics
atomistic simulation of condensed matter. Molecular vibration must
be accommodated. Then computations of heroic effort are required
to explore many orders of magnitude above
it.

Figure 10.2: Example of a crystal lattice atomistic simulation in three
dimensions. Study of a region of nanocrystaline metal with 840,000
atoms ready to be deformed. (Courtesy: Ju Li, Massachusetts
Institute of Technology.)
| 1. | Calculate the force at current position x on each particle
due to all the others. |
| 2. | Accelerate and move particles for ∆t, getting
new velocities v and positions x. |
| 3. | Repeat from 1.
|
Generally a fast second order accurate scheme for the acceleration and
motion (stage 2) is needed. One frequently used is the leap-frog
scheme. Another is called the Verlet scheme,
which can be expressed as
where an is the acceleration corresponding to position
xn.62
We also usually need to store a record of where the particles go,
since that is the major result of our simulation. And we need methods
to analyse and visualize the large amount of data that will result: the
number of steps Nt times the number of particles Np times at
least 6 (3 space, and 3 velocity) components.
10.1.1 Atomic/Molecular Forces and Potentials
The simplest type of forces, but still useful for a wide range of
physical situations, are particle-pair attraction and repulsion. Such
forces act along the vector r=x1−x2 between the
particle positions, and have a magnitude that depends only upon the
distance r=|x1−x2| between them. An example might be
the inverse-square electric force between two charges q1 and q2,
F = (q1q2/4πϵ0) r/r3. But for atomistic
simulation more usually neutral particles are being modelled whose
force changes from mutual attraction at longer distances to mutual
repulsion at short distances.
 (a)
(a)
 (b)
(b)
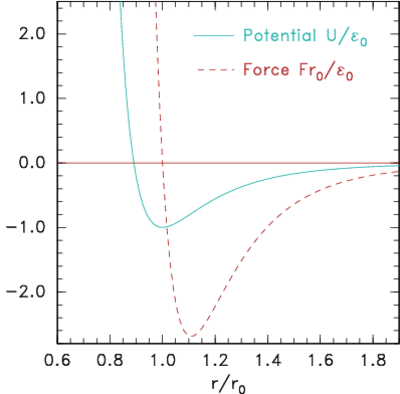
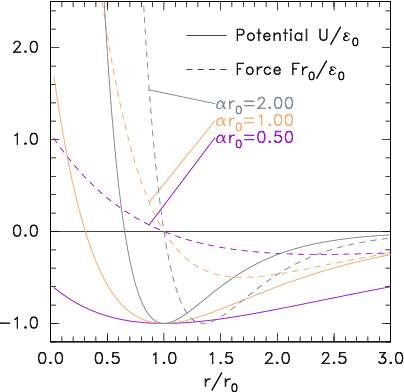
Figure 10.3: Potential and corresponding force forms for (a)
Lennard-Jones (eq.
10.2) and (b) Morse (eq.
10.3) expressions.
|
F=−∇U with U = E0 | ⎡
⎣
| ⎛
⎝
|
r0
r
| ⎞
⎠
|
12
|
−2 | ⎛
⎝
|
r0
r
| ⎞
⎠
|
6
| ⎤
⎦
|
|
| (10.2) |
A simplicity, but also a weakness, in the Lennard-Jones form is that
it depends upon just two parameters, the typical energy E0,
and the typical distance r0. The equilibrium distance,
corresponding to where the force (−dU/dr) is zero is, is r0. At
this spacing the binding energy is E0. The maximum
attractive force occurs where d2U/dr2=0, which is r=1.109 r0
and it has magnitude 2.69E0/r0. The weakness of having only
two parameters is that the spring-constant for the force, d2U/dr2,
near the the equilibrium position cannot be set independent of the
binding energy. An alternative force expression that allows this
independence, by having three parameters, is the
Morse form
|
U = E0 (e−2α(r−r0)−2 e−α(r−r0)). |
| (10.3) |
Its force is zero at r0, where the binding energy is E0;
but the spring constant there can be adjusted using the parameter
α.
See Fig. 10.3(b).
These simple two-particle, radial-force, forms omit several phenomena
that are important in molecular interactions in nature. These
additional phenomena include higher order potential interactions,
represented by the total potential energy of the entire assembly being given
as a heirarchy of sums of multiple-particle interactions
|
U = |
∑
i
|
U1(xi) + |
∑
ij
|
U2(xi,xj) + |
∑
ijk
|
U3(xi,xj,xk) + ... |
| (10.4) |
where the subscripts i,j,... refer to different particles.
The force on a particular particle l is then
. The first term U1 represents a
background force field. The second represents the pairwise force that
we've so far been discussing. We've been considering the particular
case where U2 depends only on r=|xi−xj|. The third
(and higher) terms represent possible multi-particle correlation
forces. They are often called "cluster" potential
terms.
Other force laws between multi-atom molecules might include the
orientation of the molecule bonds. In that case, internal orientation
parameters would have to be introduced or else the molecule's
individual atoms themselves represented by particles whose bonds are
modelled using force laws appropriate to them. They could be
represented as a third or probably at least fourth order series for
the potential.
10.1.2 Computational requirements
If there are Np particles, then to evaluate
the force on particle i from all the other particles j, requires
Np force evaluations for pair-forces (U2 term). It requires
Np2 for three-particle (U3) terms and so on. The force
calculation needs to be done for all the particles at each step, so if
we include even just the pair-forces for all particles, Np2 force
terms must be evaluated. This is too much. For example, a million
particles would require 1012 pair force evaluations per time
step. Computational resources would be
overwhelmed.
Therefore the most important enabling
simplification of a practical atomistic simulation is to reduce the
number of force evaluations till it is not much worse than linear in
Np. This can be done by taking advantage of the fact that the force
laws between neutral atoms have a rather
short range;
so the forces can be ignored for particle spacings greater
than some modest length. In reality we only need to calculate the
force contributions from a moderately small number of nearby particles
on each particle i. It is not sufficient to look at the position of
all the other particles at each step and decide whether they are near
enough to worry about. That decision is itself an order Np cost per
particle (Np2 total). Even if it's a bit cheaper than actually
evaluating the force, it won't do. Instead we have to keep track, in
some possibly approximate way, of which of the other particles are
close enough to the particle i to matter to it.
There are broadly two ways of doing this. Either we literally keep a
list of near neighbors associated with each
particle. Or else we divide up the volume under consideration into
much smaller blocks and adopt the policy of only examining the
particles in its own block and the neighboring
blocks. Either of these will obviously work for
a crystal lattice type
problem, modelling a solid, because the atoms hardly ever change their
nearest neighbors, or the members of blocks. But in liquids or gases
the particles can move sufficiently that their neighbors or blocks are
changing. To recalculate which particles are neighbors costs ∼ Np2. However, there are ways to avoid having to do the neighbor
determination every step. If we do it rarely enough, we reduce the
cost scaling.
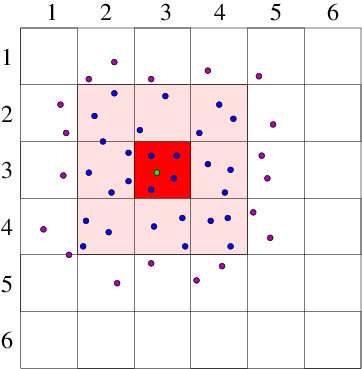
Neighbor List Algorithm
A common way to maintain an
adequately accurate neighbor list is as follows. See Fig. 10.4.
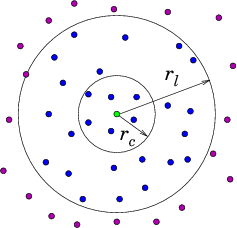 (a)
(a)
 (b)
(b)
Figure 10.4: Neighbor List (a), and Block (b), algorithms enable testing
only nearby particles for force influence. The neigbors whose
influence must be calculated are inside the radius r
l or inside
the shaded region.
10.2 Particle in Cell Codes
If the interparticle force law is of infinite
range, as it is, for example, with the
inverse-square interactions of
charged particles in a plasma, or gravitating stars, then the
near-neighbor reduction of the force calculation does not work,
because there is no cutoff radius beyond which the interaction is
negligible. This problem is solved in a different way, by representing
the long-range interactions as the potential on a mesh of cells. This
approach is called "Particle in Cell" or PIC64.
Consider, for simplicity, a single species of charged particles (call
them electrons) of charge q (=−e for electrons, of
course), and mass m. Positive ions could also be modelled
as particles, but for now take them to be represented by a smooth
neutralizing background of positive charge density −niq. The
electrons move in a region of space divided into cells labelled with
index j at positions xj. [Most modern PIC production codes are
multidimensional, but the explanations are easier in one dimension.]
They give rise to an electric potential ϕ. Ignoring the
discreteness of the individual electrons, there is a smoothed-out
total (sum of electron and ion) charge density ρq(x) = q[n(x)−ni]. The potential satisfies Poisson's
equation
|
∇2ϕ = |
d2ϕ
dx2
|
= − |
ρq
ϵ0
|
= − |
q[n(x)−ni]
ϵ0
|
|
| (10.5) |
We represent this potential discretely on the mesh: ϕi and solve
it numerically using a standard elliptic solver. The only new
feature is that we need to obtain a measure of smoothed-out density on
the mesh. We do this by assigning the charge density of the individual
electrons to the mesh in a systematic way. The simplest way to assign
it is to say
that each electron's charge is assigned
to the Nearest Grid Point NGP.
That's equivalent
to saying each electron is like a rod of length equal to the distance
∆x between the grid points, and it contributes charge density
equal to q/∆x from all positions along its length. See Fig. 10.5. The volume
of the cell is ∆x and the electron density is equal to the
number of particles whose charge is assigned to that cell divided by
the cell volume.
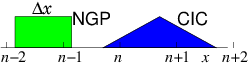
Figure 10.5: Effective shapes for the NGP and CIC charge assignments.
| 1. | Assign the charge from all the particles onto the grid cells. |
| 2. | Solve Poisson's equation to find the potential ϕj. |
| 3. | For each particle, find ∇ϕ at its position, xi, by
interpolation from the xj. |
| 4. | Accelerate, by the corresponding force, and move the particles. |
| 5. | Repeat from 1. |
This process will simulate the behavior of the plasma accounting
realistically for the particle motion. So it is a kind of atomistic
simulation. Fig. 10.6 shows an illustration of an
example in a spherical computational domain.

Figure 10.6: A curved grid (relatively unusual for PIC) shaded to
represent the density normalized to the distant value.
A few representative particle orbits in the vicinity of a spherical
object are shown, and arrows indicate the mean ion
velocity.
10.2.1 Boltzmann Equation Pseudoparticle Representation
In a particle in cell code, the particles move and are tracked in
phase-space: (x,v) is known at each time-step. A particle's
equation of motion in phase space is
|
|
d
dt
|
| ⎛
⎝
|
x
v
| ⎞
⎠
|
= | ⎛
⎝
|
v
a
| ⎞
⎠
|
. |
| (10.6) |
This is also the equation of motion of the characteristics of
the Boltzmann equation (8.12,8.13). Thus
advancing a PIC code with Np particles is equivalent to integrating
along Np characteristics of the Boltzmann equation. But what about
collisions?
The remarkable thing about a PIC code, in its simplest implementation,
is that it has essentially removed all charged-particle
collisions. The grainy discreteness of the electrons, is smoothed away
by the process of assigning charge to the grid and then solving for
ϕ. Therefore, unless we do something to put collisions back, the
PIC code actually represents integration along characteristics of the
Vlasov equation, the
collisionless Boltzmann equation. If we had instead used
(highly inefficiently) pair-forces, then the charged particle
collisions would have been retained.
Because the collisions have been removed from the problem, the actual
magnitude of each particle's charge and mass no longer matters; only
the ratio, q/m, appears in the acceleration, a, in
the Vlasov equation. That means we can represent physical situations
that would in nature involve unmanagable numbers of physical electrons by
regarding the electrons (or ions) of our computation as
pseudo-particles. Each pseudo-particle corresponds to a very
large number of actual particles, reducing the number of computation
pseudo-particles to a managable total, and keeping the costs of the
computation within tolerable bounds. For our computation to remain a
faithful representation of the physical situation, we require only
that the resolution in phase-space, which depends upon the total
number of randomly distributed electrons, should be sufficient for whatever
phenomenon is being studied. We also require, of course, that the
potential mesh has sufficient spatial resolution.
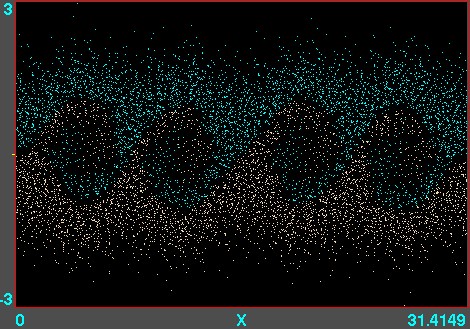
Figure 10.7: Example of phase-space locations of electrons. A
one-dimensional v versus x calculation is illustrated, using
the code XES1 (by Birdsall, Langdon, Verboncoeur and Vahedi,
originally distributed with the text by Birdsall and Langdon) for
two streams of particles giving rise to an instability whose
wavelength is four times the domain length. Each electron position
is marked as a point. Their motion can be viewed like a
movie.
10.2.2 Direct Simulation Monte Carlo treatment of gas
An approach that combines some of the features of PIC and atomistic
simulation, is the treatment of tenuous neutral gas behavior by what
have come to be called Direct Simulation Monte Carlo
(DSMC) codes.
These are for addressing situations where the ratio of the
mean-free-path of molecules to the
characteristic spatial feature size
(the "Knudsen number") is of order unity
(within a factor of 100 or so either way). Such situations occur in
very tenuous gases (e.g. orbital re-entry in space) or when the
features are microscopic. DSMC shares with PIC the features that the
domain is divided into a large number of cells, that
pseudo-particles
are used, and that collisions are represented in simplified way that
reduces computational cost and yet approximates physical
behavior. DMSC is also, in effect, integrating the Boltzmann equation
along characteristics, but in this case there's no acceleration term,
so the characteristics are straight lines.
The pseudo-particles representing molecules are advanced in time, but
at each step, chosen somewhat shorter than a typical collision time,
they are examined to decide whether they have collided. In order to
avoid a Np2 cost, collisions are considered only with the
particles in the same cell of the grid. (This partitioning is all the
cells are used for.) The cells are chosen to have size smaller than a
mean-free-path, but not by much. They will generally have only a
modest number (perhaps 20-40) of pseudo-particles in each cell. The
number of individual molecules represented by each pseudo-particle is
adjusted to achieve this number per cell.
Whether a collision has occurred between two particles is decided
based only upon their relative velocity, not on their position
within the cell. This is the big approximation. A statistical test
using random numbers decides if and which collisions have happened. A
collision changes the velocity of both colliding particles, in
accordance with the statistics of the collision cross-section and
corresponding kinematics. That way, momentum and energy are
appropriately conserved within the cell as a whole. Steps are
iterated, and the overall behavior of the assembly of particles is
monitored and analysed to provide effective fluid parameters like
density, velocity, effective viscosity, and so on. Fig. 10.8
shows an example from the code DSMC, v3.0 developed by Graeme Bird.
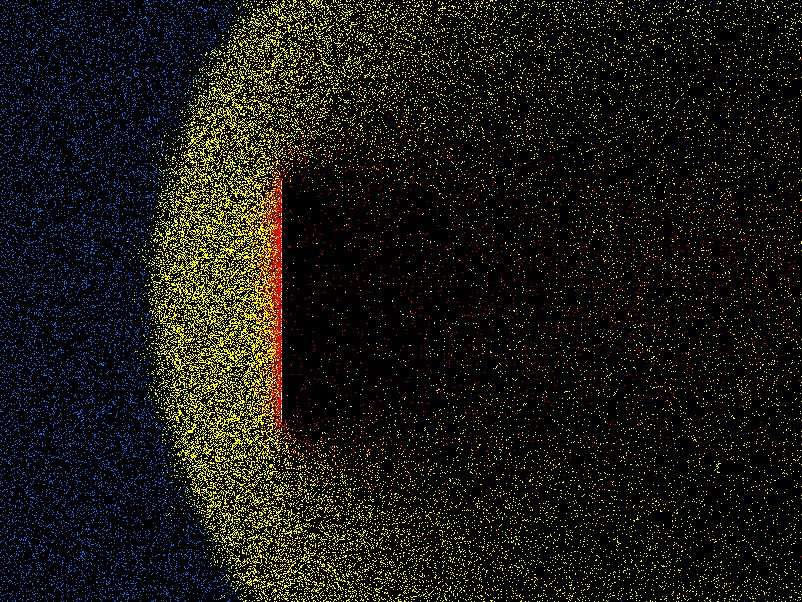
Figure 10.8: Example of position plot in two space dimensions of tenuous
gas flow past a plate. Different colors (shadings) indicate molecules that
have been influenced by the plate through collisions.
10.2.3 Particle Boundary Conditions
Objects that are embedded in a particle computation region present
physical boundaries at which appropriate approximate conditions must
be applied. For example with DSMC, gas particles are usually reflected,
whereas with plasmas, it is usually assumed that the electrons are removed
by neutralization when they encounter a solid surface.
An important question arises in most particle simulation methods. What
do we do at the outer boundary of our computational domain? If a particle
leaves the domain, what happens to it? And what do we do to represent
particles entering the domain?
Occasionally the boundary of our domain might be a physical boundary
no different from an embedded object. But far more often the edge of
the domain is simply the place where our computation stops, not where
there is any specific physical change. What do we do then?
The appropriate answer depends upon the specifics of the situation,
but quite often it makes sense to use periodic boundary
conditions. Periodic conditions
for particles are like periodic conditions for differential equations
discussed in section 3.3.2. They treat the particles as if a
boundary that they cross corresponds to the same position in space as
the opposite boundary. A particle moving on a computational domain in
x that extends from 0 to L, when it steps past L, to a new
place that would have been x=L+δ, outside the domain, is
reinserted at the position x=δ close to the opposite boundary,
but back inside the domain. Of course the particle's velocity is just
what it would have been anyway. Velocity is not affected by the
reinsertion process. Periodic conditions can be applied in any number
of dimensions.
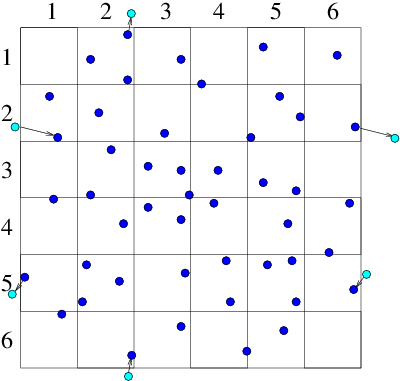 (a)
(a) (b)
(b)
Figure 10.9: Particles that cross outer periodic boundaries (a) are
relocated to the opposite side of the domain. This is equivalent (b)
to modelling an (infinite) periodic array made up of repetitions
of the smaller domain.
Worked Example: Required resolution of PIC grid
How fine must the potential mesh be in an electron Particle in Cell code?
Well, it depends on how fine-scale the potential variation might
be. That depends on the parameters of the particles
(electrons). Suppose they have approximately a Maxwell-Boltzmann
velocity-distribution of temperature Te. We can estimate the finest
scale of potential variation as follows. We'll consider just a
one-dimensional problem. Suppose there is at some position x, a
perturbed potential ϕ(x) such that |ϕ| << Te/e, measured
from a chosen reference ϕ∞=0 in the equilibrium background
where the density is ne=n∞=ni. (Referring to the
background as ∞ is a helpful notation that avoids implying the
value at x=0; it means the distant value.) Then the electron
density at x can be deduced from the fact that f(v) is constant along
orbits (characteristics). In the collisionless steady-state,
energy is conserved; so for any orbit 1/2mv2−eϕ = 1/2mv∞2, where v∞ is the velocity on that orbit when it
is "at infinity" in the background, where ϕ = 0. Consequently,
| f(v)=f∞(v∞) = n∞ |
⎛
√
|
|
exp(−mv∞2/2Te) = n∞ |
⎛
√
|
|
exp(−mv2/2Te+eϕ/Te)
|
. Hence at x, f(v) is Maxwellian with density
n=∫f(v)dv=n∞exp(eϕ/Te).
Now let's find analytically the steady potential arising for x > 0 when the
potential slope at x=0 is dϕ/dx=−E0. Poisson's equation in
one-dimension is
|
|
d2ϕ
dx2
|
= − |
en∞
ϵ0
|
[1−exp(eϕ/Te)] ≈ | ⎛
⎝
|
e2n∞
ϵ0 Te
| ⎞
⎠
|
ϕ. |
| (10.7) |
The final approximate form gives Helmholtz's equation. It is obtained by
Taylor expansion of the exponential to first order, since its argument
is small. The solution satisfying the condition at x=0 is then
|
ϕ(x)=E0λe−x/λ, where λ2 = | ⎛
⎝
|
e2n∞
ϵ0 Te
| ⎞
⎠
|
. |
| (10.8) |
Based on this model calculation, the length
, which is called the Debye
length, is the characteristic spatial scale of
potential variation. A PIC calculation must have fine enough grid to
resolve the smaller of λ and the characteristic feature-size
of any object in the problem whose influence introduces potential
structure. In short, ∆x ≤ λ.
If we had an operational PIC code, we could do a series of
calculations with different cell size ∆x. We would find that
when ∆x became small enough, the solutions would give a result
independent of ∆x. That would be a good way of demonstrating
adequate spatial resolution numerically. For
the simple problem we've considered the requirement can be calculated
analytically. Actually the criterion ∆x <~λ applies
very widely in plasma PIC calculations.
Exercise 10. Atomistic Simulation
1. The Verlet scheme for particle advance is
Suppose that the velocity at integer timesteps is related to that at
half integer timesteps by
vn=(vn−1/2+vn+1/2)/2.
With this identification, derive the Verlet scheme from
the leap-frog scheme,
and thus show they are equivalent.
2. A block algorithm is applied to an atomistic
simulation in a cubical 3 dimensional region, containing Np=1000000
atoms approximately uniformly distributed. Only two-particle forces
are to be considered. The cut-off range for particle-particle force is
4 times the average particle spacing. Find
(a) The optimal size of blocks into which to divide the domain for
fastest execution.
(b) How many force evaluations per time-step will be required.
(c) If the force evaluations require 5 multiplications, a Verlet
advance is used, and the calculation is done on a single processor
which takes 1 nanosecond per multiplication on average, roughly what
is the total time taken per timestep (for the full 1,000,000 particles).
3. (a) Prove from the definition of a characteristic
(see section 8.3.2) that the equation of the
characteristics of the collisionless Boltzmann equation is
|
|
d
dt
|
| ⎛
⎝
|
x
v
| ⎞
⎠
|
= | ⎛
⎝
|
v
a
| ⎞
⎠
|
. |
| (10.11) |
Show also that a particle (or pseudo-particle) trajectory in a fixed
electric potential depends only on initial velocity and the ratio of
its charge to mass q/m, and therefore that the equation of motion of
a pseudo-particle must use the same q/m as the actual particles, if
a PIC simulation is to model correctly the force law at given speed
and potential.
(b) A pseudo-particle of charge q follows a characteristic, but it
is supposed to be representative of many nearby particles
(characteristics). If the mean density of pseudo-particles in a PIC
simulation is a factor 1/g (where g >> 1) smaller than the actual
density of the system being modelled, how much charge must each
pseudo-particle deposit on the potential grid to give the correct
potential from Poisson's equation? One way to do PIC simulation is to
represent all lengths, times, charges and masses in physical units,
but to use this charge deposition factor, and correspondingly lower
particle density.


 (a)
(a)
 (b)
(b)
 (a)
(a)
 (b)
(b)
 Usually a continuous linear
interpolation is preferred, called the Cloud In
Cell CIC assignment. The
charge density assigned from each electron is equal to q/∆x when
the electron is exactly at xj, and falls linearly, reaching zero when
the particle is at xj±1. Thus the electron is like a rod of length
2∆x whose charge distribution is triangular.
The way the particle in cell code runs is this.
Usually a continuous linear
interpolation is preferred, called the Cloud In
Cell CIC assignment. The
charge density assigned from each electron is equal to q/∆x when
the electron is exactly at xj, and falls linearly, reaching zero when
the particle is at xj±1. Thus the electron is like a rod of length
2∆x whose charge distribution is triangular.
The way the particle in cell code runs is this.



 (a)
(a) (b)
(b)